Chapter 5 Risk Ratio
5.1 はじめに
Davis and Weller (2021) の論文からデータを用いる。
スタチン療法は多くの循環器系の疾患を減少させることが知られている。一方で、スタチン関連の筋症状のためアドヒアランスが低く、結果として循環器系アウトカムの低下に繋がっている可能性がある。さらに、スタチンの量とも関係していることが予想されるが、量の多い (high intensity) スタチン療法の研究は少ない。そのため、ネットワークメタ分析で、placebo-moderate, moderato-high, placebo-high の比較を行った。
5.2 頻度論 netmeta
5.2.1 データ
arm-based から読み込む場合は、以下のようになる。
dfNetMeta <- pairwise(
treat = treat,
event = X,
n = Total,
studlab = Study,
data = dfArm,
sm ="RR")netmetaDavis <- netmeta(TE, seTE, treat1, treat2, studlab, data = dfNetMeta, sm="RR", reference="Placebo")contrast-based データの場合は、以下のようになる。なお、treat1 = treat1 の左側の treat1 は、この関数の引数名である。右側の treat1 は、エクセル中の列名(データフレームの列名)である。
netmetaShim <- netmetabin(
treat1 = treat1,
treat2 = treat2,
n1 = n1,
n2 = n2,
event1 = event1,
event2 = event2,
studlab = study,
data=dfCon,
sm="RR",
reference="Placebo")Xie 2016 が削除された。おそらくイベント数が0のものがあるためだろうか?arm-based データを pairwise() で計算すると、event = 0 を +0.5 して OR を計算するので、arm-based の方が良いかもしれない。
5.2.2 Network plot
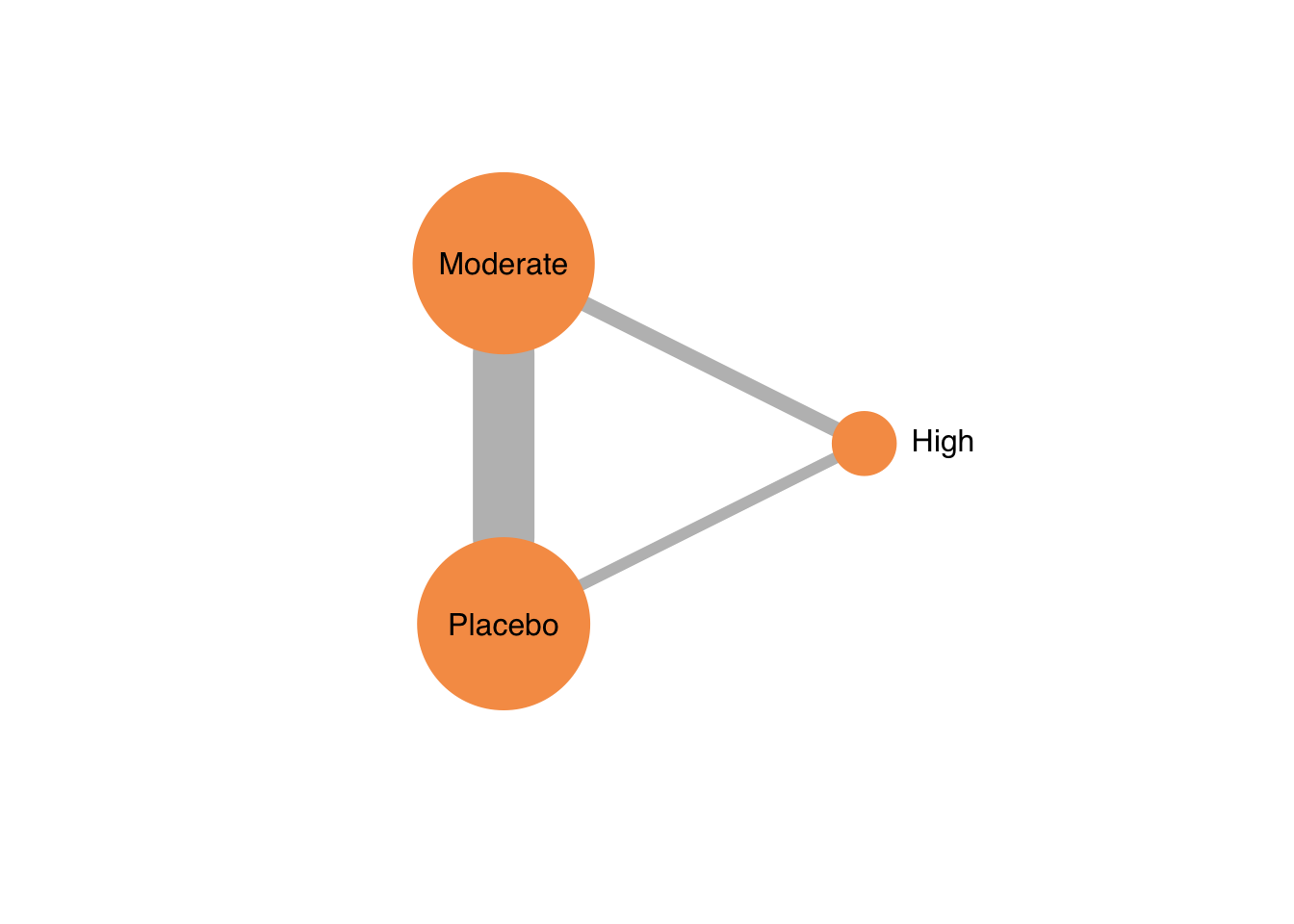
netgraph(netmetaDavis,
plastic = FALSE, # 3Dではなくする
points = TRUE, # ノードを表示する
thickness = "number.of.studies", # 線の太さを研究数にする
multiarm = TRUE) # multiarm のところを塗りつぶす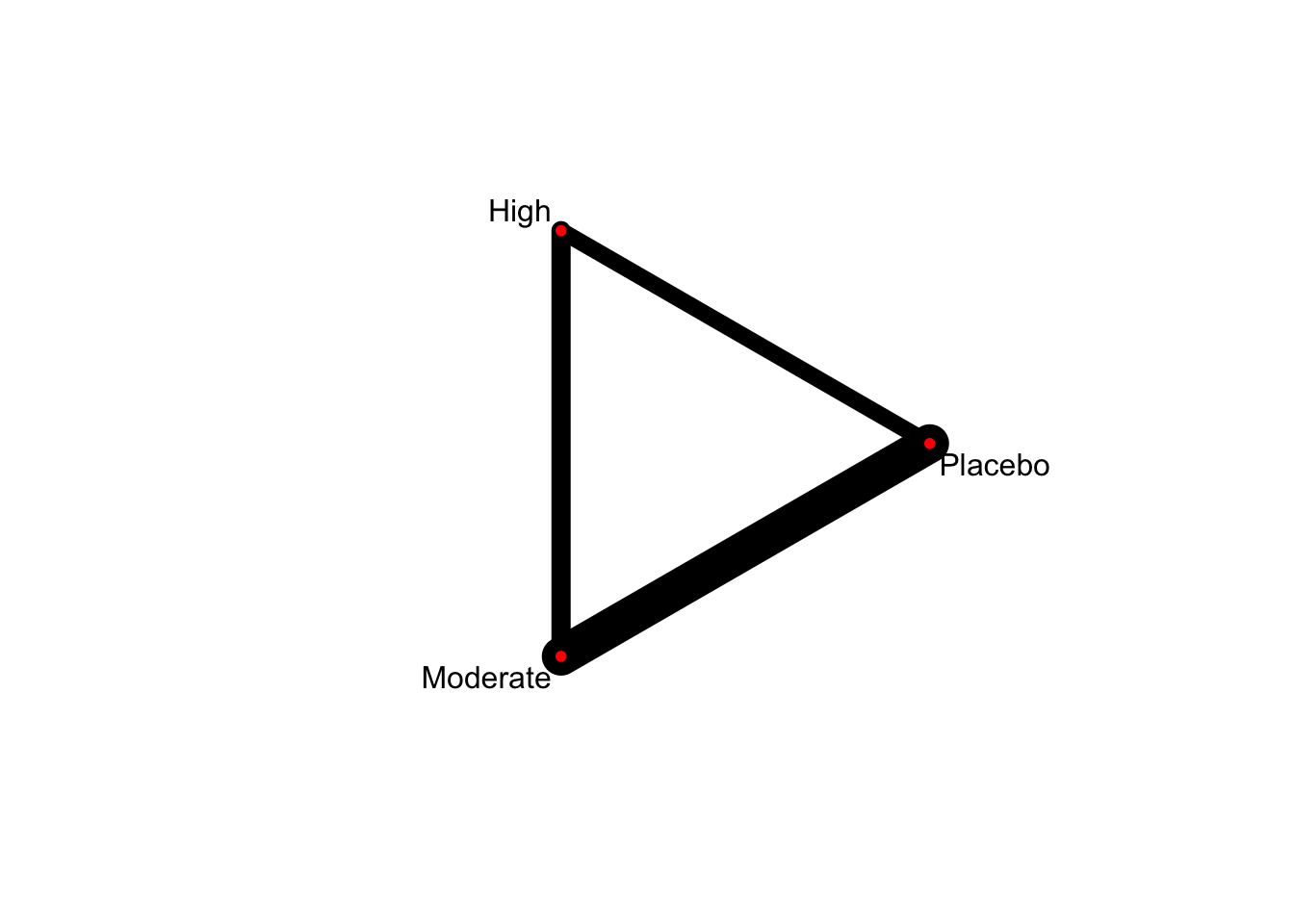
ノードの大きさを変更することはできない。
5.2.3 要約
## Original data:
##
## treat1 treat2 TE seTE
## 4D, A20 Moderate Placebo 0.3636 0.5828
## A to Z, S40-S80 vs S20 High Moderate 0.1725 0.2300
## AFCAPS, L20-L40 Moderate Placebo 0.0399 0.0197
## ALERT, F40-F80 Moderate Placebo -0.0056 0.0433
## ASCOT, A10 Moderate Placebo 0.0473 0.0806
## ASPEN, A10 Moderate Placebo 0.6291 0.2806
## AURORA, R10 Moderate Placebo -0.1091 0.0685
## CARDS, A10 Moderate Placebo -0.0167 0.0512
## CARE, P40 Moderate Placebo -2.1958 1.4904
## CORONA,R10 Moderate Placebo 0.0766 0.0921
## GISSI-HF, R10 Moderate Placebo 0.0927 0.3004
## HOPE, R10 Moderate Placebo 1.0945 1.1544
## HPS, S40 Moderate Placebo -0.0090 0.0199
## JUPITER, R20 High Placebo 0.0323 0.0347
## LIPID, P40 Moderate Placebo -0.2254 0.4739
## PROSPER, P40 Moderate Placebo 0.1254 0.2415
## PROVE-IT, A80 vs P40 High Moderate 0.1915 0.1772
## SEARCH, S80 vs S20 High Moderate 0.0428 0.0212
## SPARCL, A80 High Placebo -0.0885 0.1183
## SSSS, S20-S40 Moderate Placebo 0.0358 0.0492
## TNT, A80 vs A10 High Moderate 0.0317 0.0896
## TRACE RA, A40 High Placebo 0.1166 0.1216
## WOSCOPS, P40 Moderate Placebo -0.0530 0.1397
##
## Number of treatment arms (by study):
## narms
## 4D, A20 2
## A to Z, S40-S80 vs S20 2
## AFCAPS, L20-L40 2
## ALERT, F40-F80 2
## ASCOT, A10 2
## ASPEN, A10 2
## AURORA, R10 2
## CARDS, A10 2
## CARE, P40 2
## CORONA,R10 2
## GISSI-HF, R10 2
## HOPE, R10 2
## HPS, S40 2
## JUPITER, R20 2
## LIPID, P40 2
## PROSPER, P40 2
## PROVE-IT, A80 vs P40 2
## SEARCH, S80 vs S20 2
## SPARCL, A80 2
## SSSS, S20-S40 2
## TNT, A80 vs A10 2
## TRACE RA, A40 2
## WOSCOPS, P40 2
##
## Results (common effects model):
##
## treat1 treat2 RR 95%-CI Q leverage
## 4D, A20 Moderate Placebo 1.0098 [0.9875; 1.0326] 0.37 0.00
## A to Z, S40-S80 vs S20 High Moderate 1.0386 [1.0035; 1.0749] 0.34 0.01
## AFCAPS, L20-L40 Moderate Placebo 1.0098 [0.9875; 1.0326] 2.33 0.33
## ALERT, F40-F80 Moderate Placebo 1.0098 [0.9875; 1.0326] 0.13 0.07
## ASCOT, A10 Moderate Placebo 1.0098 [0.9875; 1.0326] 0.22 0.02
## ASPEN, A10 Moderate Placebo 1.0098 [0.9875; 1.0326] 4.87 0.00
## AURORA, R10 Moderate Placebo 1.0098 [0.9875; 1.0326] 3.01 0.03
## CARDS, A10 Moderate Placebo 1.0098 [0.9875; 1.0326] 0.27 0.05
## CARE, P40 Moderate Placebo 1.0098 [0.9875; 1.0326] 2.19 0.00
## CORONA,R10 Moderate Placebo 1.0098 [0.9875; 1.0326] 0.53 0.02
## GISSI-HF, R10 Moderate Placebo 1.0098 [0.9875; 1.0326] 0.08 0.00
## HOPE, R10 Moderate Placebo 1.0098 [0.9875; 1.0326] 0.88 0.00
## HPS, S40 Moderate Placebo 1.0098 [0.9875; 1.0326] 0.90 0.33
## JUPITER, R20 High Placebo 1.0488 [1.0104; 1.0886] 0.19 0.30
## LIPID, P40 Moderate Placebo 1.0098 [0.9875; 1.0326] 0.25 0.00
## PROSPER, P40 Moderate Placebo 1.0098 [0.9875; 1.0326] 0.23 0.00
## PROVE-IT, A80 vs P40 High Moderate 1.0386 [1.0035; 1.0749] 0.75 0.01
## SEARCH, S80 vs S20 High Moderate 1.0386 [1.0035; 1.0749] 0.05 0.68
## SPARCL, A80 High Placebo 1.0488 [1.0104; 1.0886] 1.32 0.03
## SSSS, S20-S40 Moderate Placebo 1.0098 [0.9875; 1.0326] 0.28 0.05
## TNT, A80 vs A10 High Moderate 1.0386 [1.0035; 1.0749] 0.00 0.04
## TRACE RA, A40 High Placebo 1.0488 [1.0104; 1.0886] 0.32 0.02
## WOSCOPS, P40 Moderate Placebo 1.0098 [0.9875; 1.0326] 0.20 0.01
##
## Results (random effects model):
##
## treat1 treat2 RR 95%-CI
## 4D, A20 Moderate Placebo 1.0098 [0.9875; 1.0326]
## A to Z, S40-S80 vs S20 High Moderate 1.0386 [1.0035; 1.0749]
## AFCAPS, L20-L40 Moderate Placebo 1.0098 [0.9875; 1.0326]
## ALERT, F40-F80 Moderate Placebo 1.0098 [0.9875; 1.0326]
## ASCOT, A10 Moderate Placebo 1.0098 [0.9875; 1.0326]
## ASPEN, A10 Moderate Placebo 1.0098 [0.9875; 1.0326]
## AURORA, R10 Moderate Placebo 1.0098 [0.9875; 1.0326]
## CARDS, A10 Moderate Placebo 1.0098 [0.9875; 1.0326]
## CARE, P40 Moderate Placebo 1.0098 [0.9875; 1.0326]
## CORONA,R10 Moderate Placebo 1.0098 [0.9875; 1.0326]
## GISSI-HF, R10 Moderate Placebo 1.0098 [0.9875; 1.0326]
## HOPE, R10 Moderate Placebo 1.0098 [0.9875; 1.0326]
## HPS, S40 Moderate Placebo 1.0098 [0.9875; 1.0326]
## JUPITER, R20 High Placebo 1.0488 [1.0104; 1.0886]
## LIPID, P40 Moderate Placebo 1.0098 [0.9875; 1.0326]
## PROSPER, P40 Moderate Placebo 1.0098 [0.9875; 1.0326]
## PROVE-IT, A80 vs P40 High Moderate 1.0386 [1.0035; 1.0749]
## SEARCH, S80 vs S20 High Moderate 1.0386 [1.0035; 1.0749]
## SPARCL, A80 High Placebo 1.0488 [1.0104; 1.0886]
## SSSS, S20-S40 Moderate Placebo 1.0098 [0.9875; 1.0326]
## TNT, A80 vs A10 High Moderate 1.0386 [1.0035; 1.0749]
## TRACE RA, A40 High Placebo 1.0488 [1.0104; 1.0886]
## WOSCOPS, P40 Moderate Placebo 1.0098 [0.9875; 1.0326]
##
## Number of studies: k = 23
## Number of pairwise comparisons: m = 23
## Number of treatments: n = 3
## Number of designs: d = 3
##
## Common effects model
##
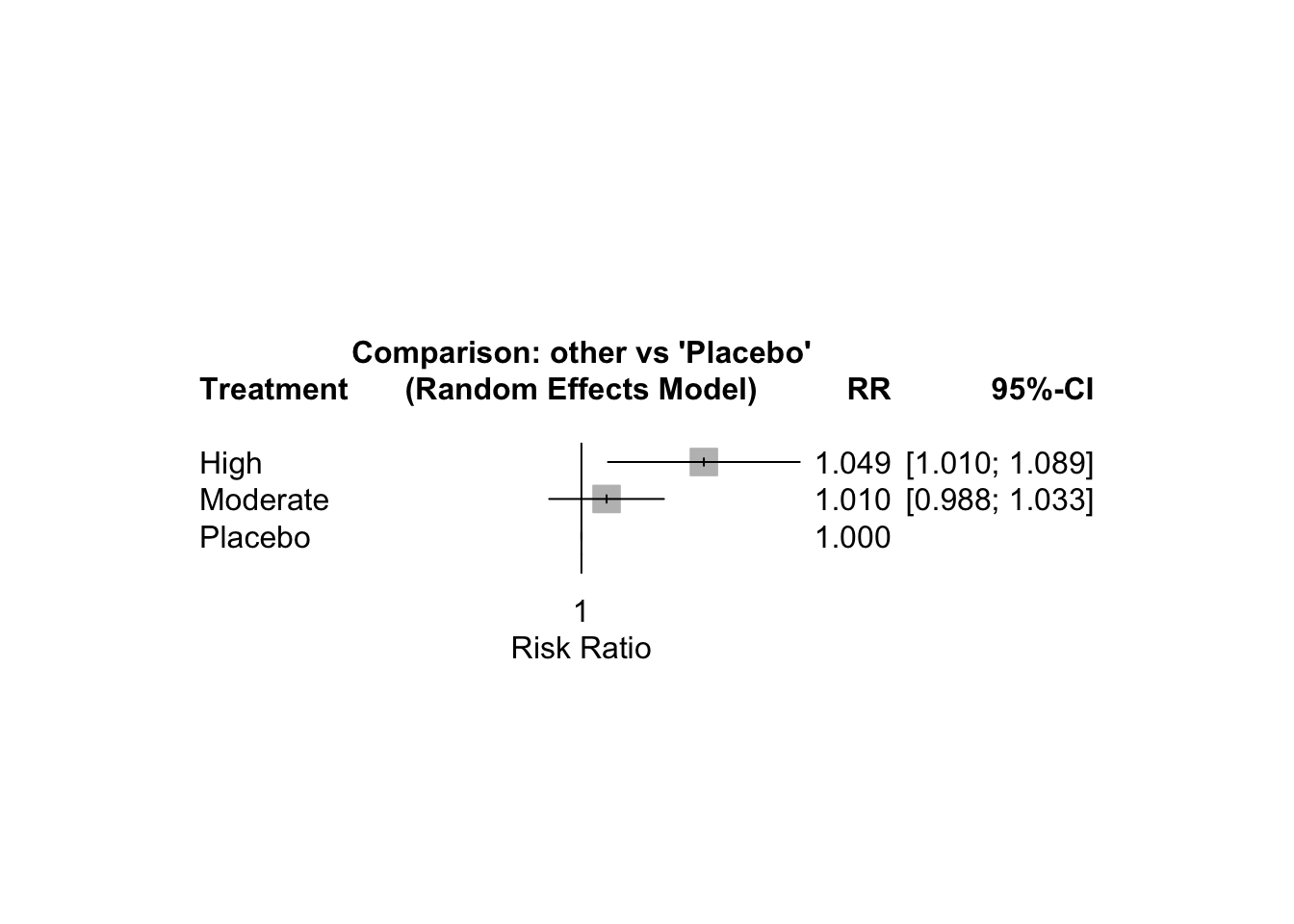
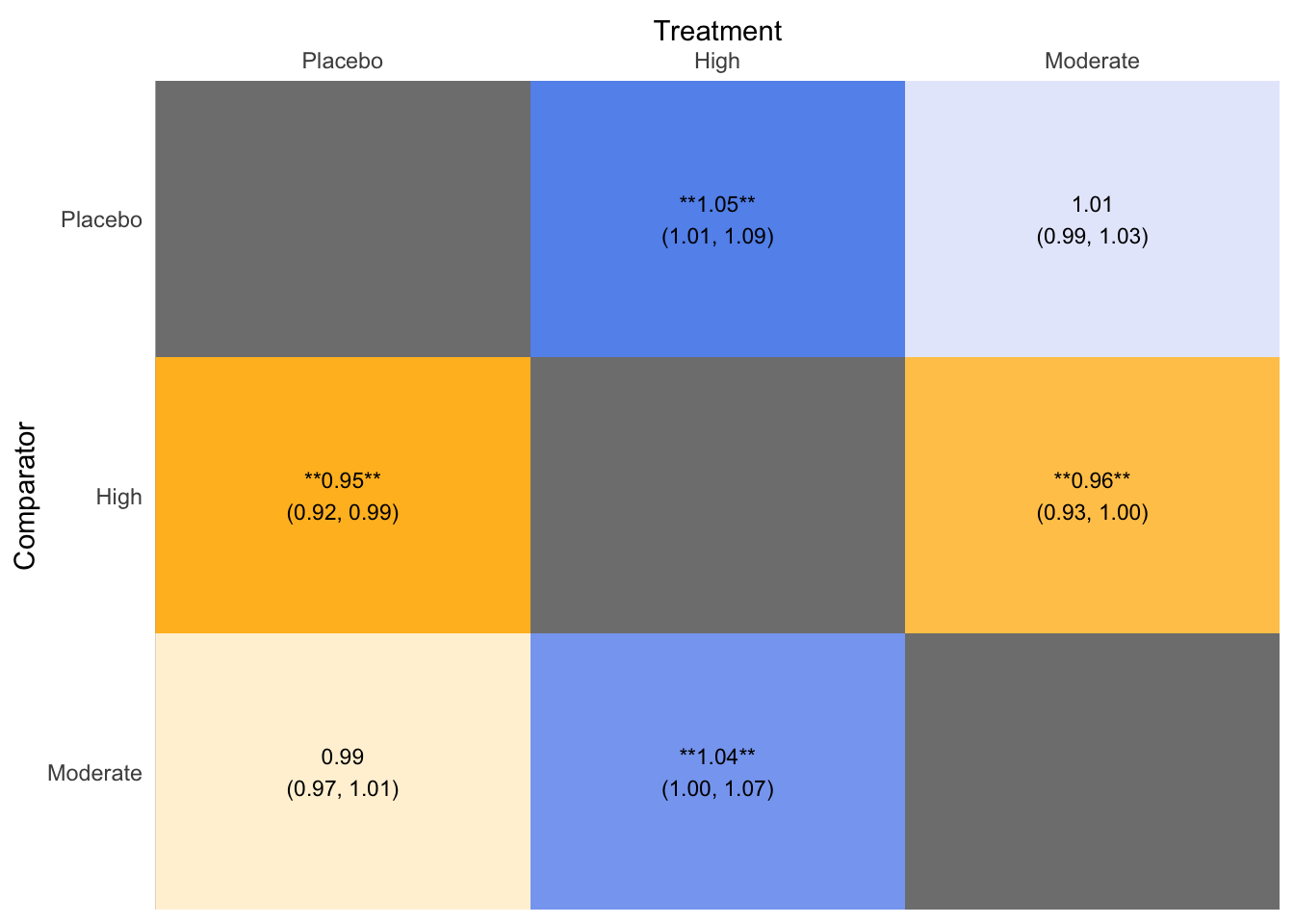
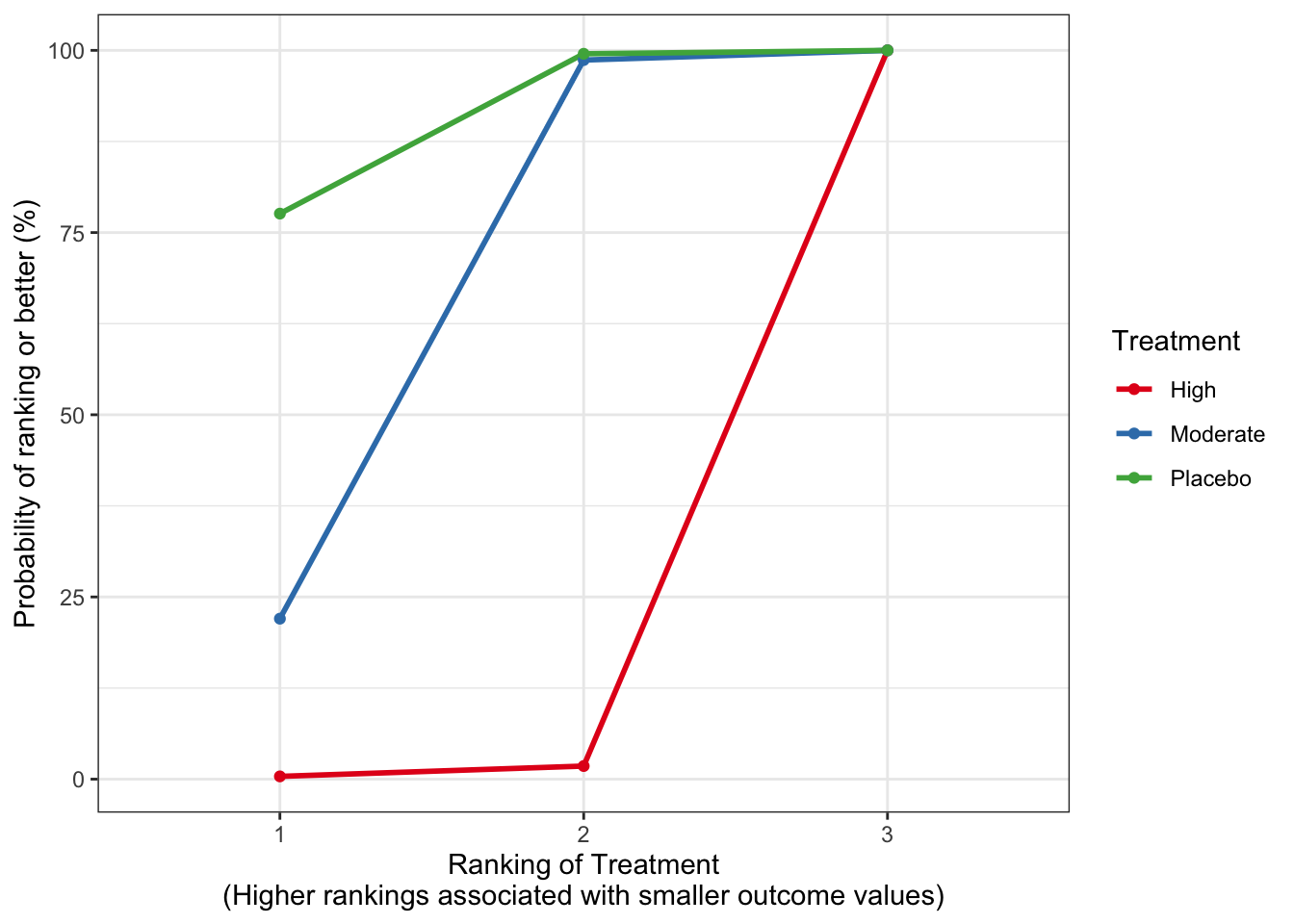
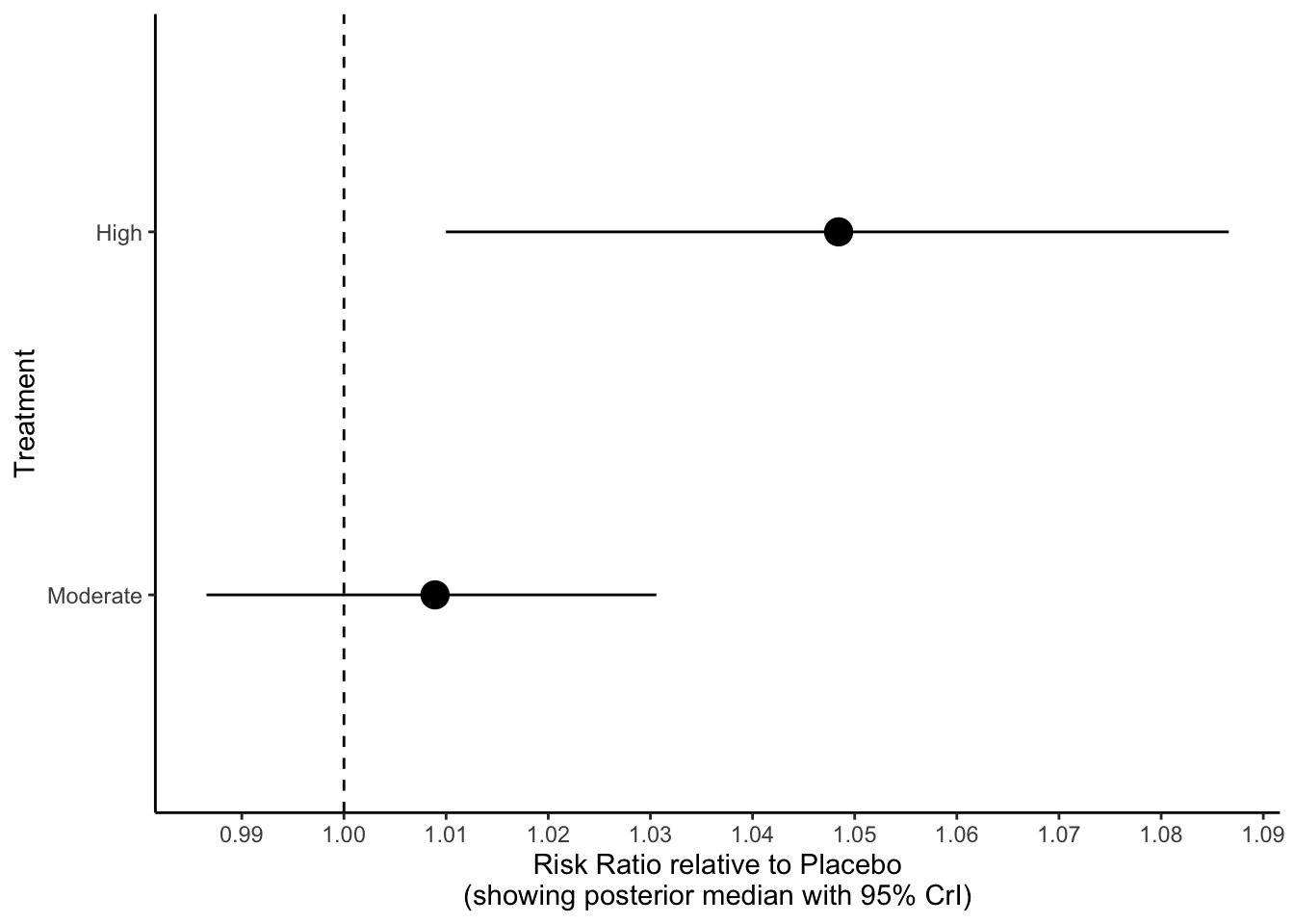
## Treatment estimate (sm = 'RR', comparison: other treatments vs 'Placebo'):
## RR 95%-CI z p-value
## High 1.0488 [1.0104; 1.0886] 2.50 0.0123
## Moderate 1.0098 [0.9875; 1.0326] 0.86 0.3904
## Placebo . . . .
##
## Random effects model
##
## Treatment estimate (sm = 'RR', comparison: other treatments vs 'Placebo'):
## RR 95%-CI z p-value
## High 1.0488 [1.0104; 1.0886] 2.50 0.0123
## Moderate 1.0098 [0.9875; 1.0326] 0.86 0.3904
## Placebo . . . .
##
## Quantifying heterogeneity / inconsistency:
## tau^2 = 0; tau = 0; I^2 = 0% [0.0%; 46.2%]
##
## Tests of heterogeneity (within designs) and inconsistency (between designs):
## Q d.f. p-value
## Total 19.71 21 0.5397
## Within designs 19.21 20 0.5082
## Between designs 0.50 1 0.47935.2.4 一貫性の評価
## Q statistics to assess homogeneity / consistency
##
## Q df p-value
## Total 4.90 19 0.9995
## Within designs 4.68 15 0.9945
## Between designs 0.22 4 0.9942
##
## Design-specific decomposition of within-designs Q statistic
##
## Design Q df p-value
## Placebo:IV(double) 1.89 7 0.9655
## Placebo:IV(single) 1.96 6 0.9229
## Placebo:Topical 0.82 2 0.6631
##
## Between-designs Q statistic after detaching of single designs
##
## Detached design Q df p-value
## IV(double):Topical 0.12 3 0.9897
## Placebo:IV(double) 0.12 3 0.9897
## Placebo:IV(single) 0.19 3 0.9798
## Placebo:Topical 0.10 3 0.9919
## Combination:IV(single):Topical 0.21 2 0.9019
## Placebo:Combination:IV(single) 0.13 2 0.9363
##
## Q statistic to assess consistency under the assumption of
## a full design-by-treatment interaction random effects model
##
## Q df p-value tau.within tau2.within
## Between designs 0.22 4 0.9942 0 0## Separate indirect from direct evidence (SIDE) using back-calculation method
##
## Common effects model:
##
## comparison k prop nma direct indir. RoR z p-value
## High:Moderate 4 0.74 1.039 1.046 1.017 1.029 0.71 0.4793
## High:Placebo 3 0.35 1.049 1.030 1.059 0.972 -0.71 0.4793
## Moderate:Placebo 16 0.91 1.010 1.012 0.984 1.029 0.71 0.4793
##
## Random effects model:
##
## comparison k prop nma direct indir. RoR z p-value
## High:Moderate 4 0.74 1.039 1.046 1.017 1.029 0.71 0.4793
## High:Placebo 3 0.35 1.049 1.030 1.059 0.972 -0.71 0.4793
## Moderate:Placebo 16 0.91 1.010 1.012 0.984 1.029 0.71 0.4793
##
## Legend:
## comparison - Treatment comparison
## k - Number of studies providing direct evidence
## prop - Direct evidence proportion
## nma - Estimated treatment effect (RR) in network meta-analysis
## direct - Estimated treatment effect (RR) derived from direct evidence
## indir. - Estimated treatment effect (RR) derived from indirect evidence
## RoR - Ratio of Ratios (direct versus indirect)
## z - z-value of test for disagreement (direct versus indirect)
## p-value - p-value of test for disagreement (direct versus indirect)p < 0.05 であると、非一貫である。
5.3 gemtc ベイジアン
gemtc は、原則として arm-based データをとる。また、gemtc は、あらかじめ列名を指定の名称にしなければならない。
library(gemtc)
dfGemtc <- dfArm
colnames(dfGemtc) <- c("study", "responders", "sampleSize", "description")
dfGemtc$treatment <- dfArm$treat
gemtcDavis <- mtc.network(data.ab=dfGemtc)5.3.1 Network plot
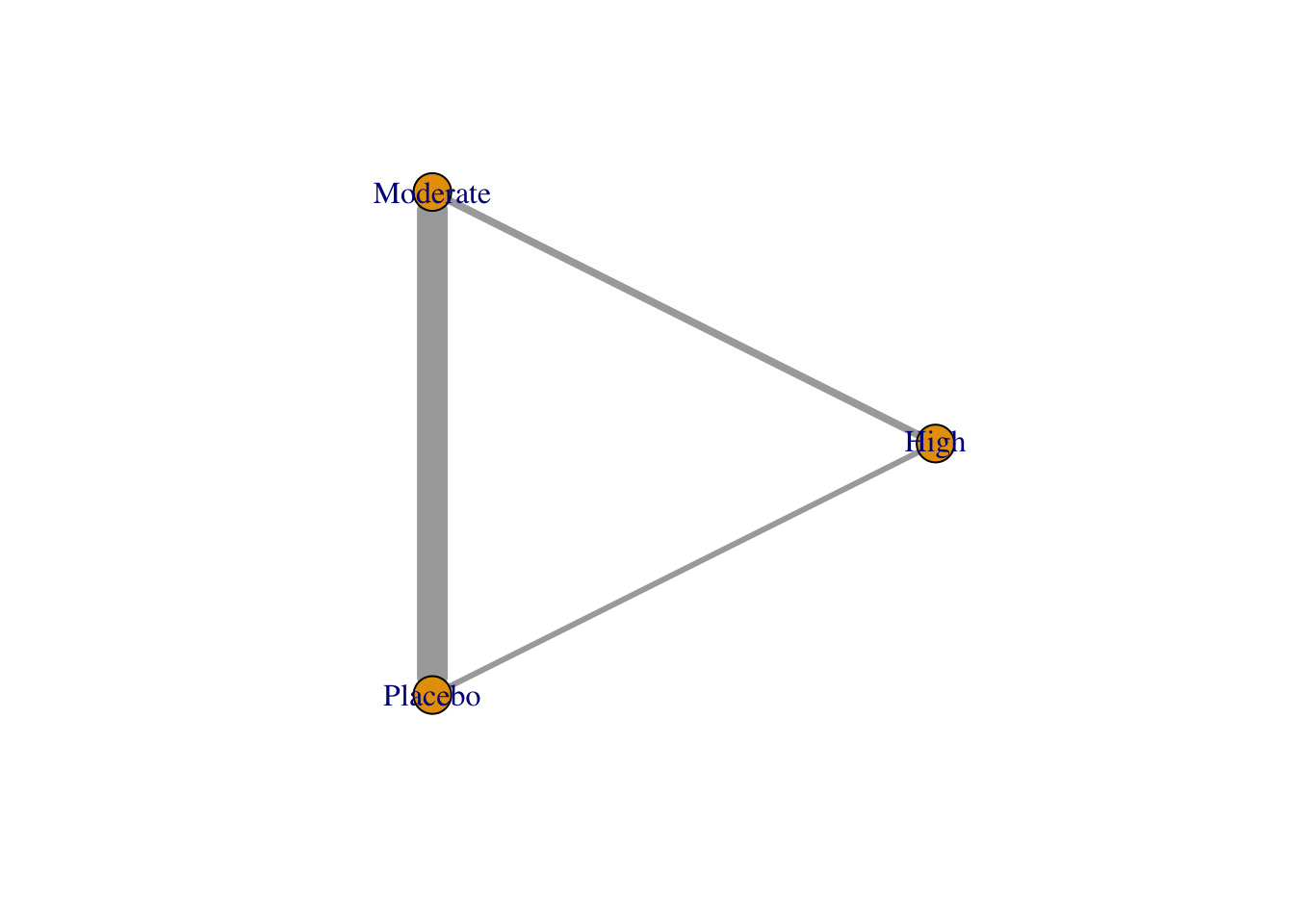
use.description で description 列を参照するはずだが、どうも機能しない。
## $Description
## [1] "MTC dataset: Network"
##
## $`Studies per treatment`
## High Moderate Placebo
## 7 20 19
##
## $`Number of n-arm studies`
## 2-arm
## 23
##
## $`Studies per treatment comparison`
## t1 t2 nr
## 1 High Moderate 4
## 2 High Placebo 3
## 3 Moderate Placebo 165.3.3 モデル実行
## Compiling model graph
## Resolving undeclared variables
## Allocating nodes
## Graph information:
## Observed stochastic nodes: 46
## Unobserved stochastic nodes: 25
## Total graph size: 932
##
## Initializing model##
## Results on the Log Odds Ratio scale
##
## Iterations = 5010:15000
## Thinning interval = 10
## Number of chains = 4
## Sample size per chain = 1000
##
## 1. Empirical mean and standard deviation for each variable,
## plus standard error of the mean:
##
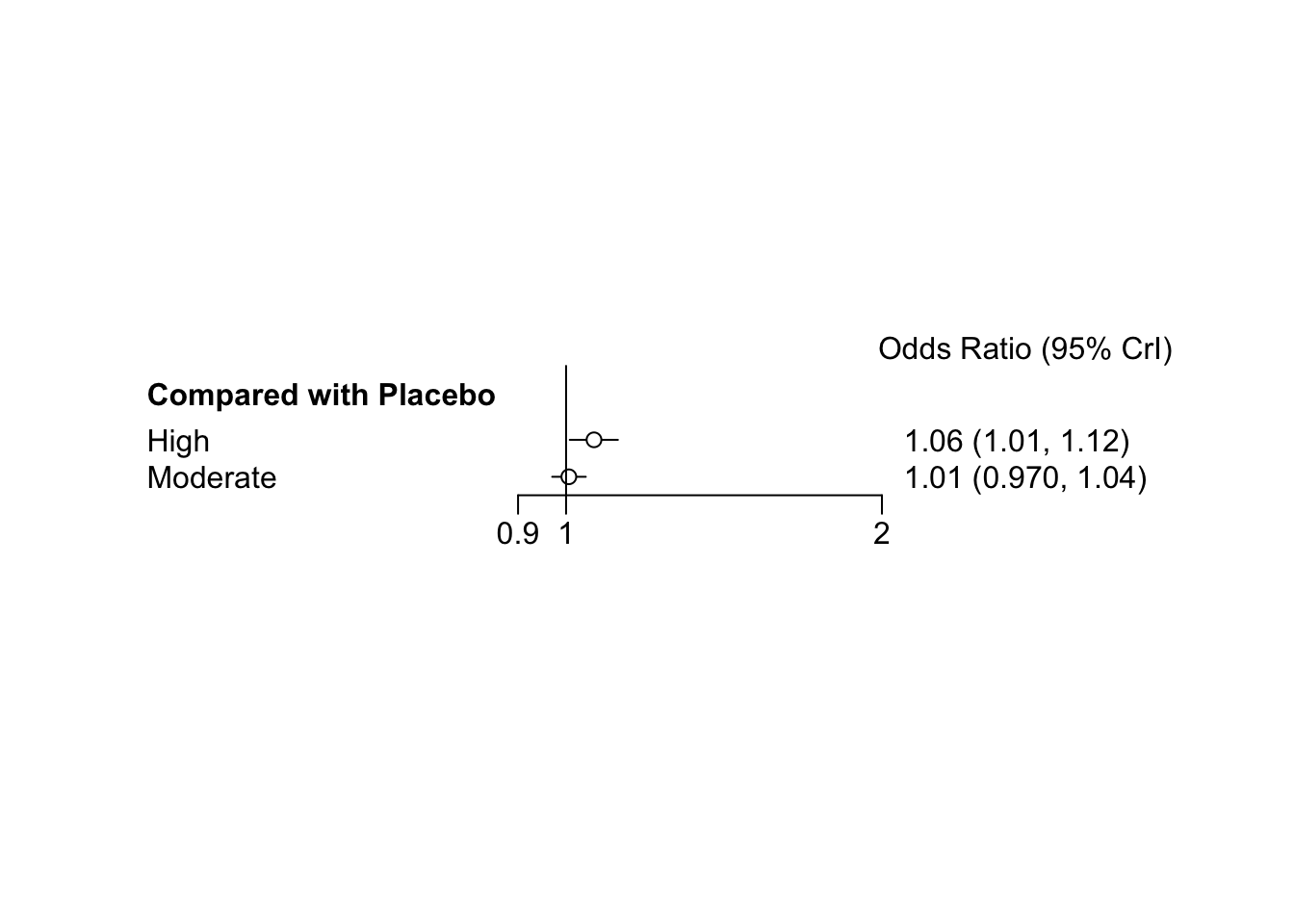
## Mean SD Naive SE Time-series SE
## d.High.Moderate -0.05467 0.02648 0.0004187 0.0005180
## d.High.Placebo -0.06094 0.02715 0.0004292 0.0005255
##
## 2. Quantiles for each variable:
##
## 2.5% 25% 50% 75% 97.5%
## d.High.Moderate -0.1063 -0.07213 -0.05463 -0.03698 -0.001840
## d.High.Placebo -0.1137 -0.07938 -0.06128 -0.04247 -0.008131
##
## -- Model fit (residual deviance):
##
## Dbar pD DIC
## 49.80489 25.31706 75.12195
##
## 46 data points, ratio 1.083, I^2 = 10%## Compiling model graph
## Resolving undeclared variables
## Allocating nodes
## Graph information:
## Observed stochastic nodes: 46
## Unobserved stochastic nodes: 49
## Total graph size: 990
##
## Initializing model##
## Results on the Log Odds Ratio scale
##
## Iterations = 5010:15000
## Thinning interval = 10
## Number of chains = 4
## Sample size per chain = 1000
##
## 1. Empirical mean and standard deviation for each variable,
## plus standard error of the mean:
##
## Mean SD Naive SE Time-series SE
## d.High.Moderate -0.05219 0.03557 0.0005624 0.001384
## d.High.Placebo -0.06220 0.03581 0.0005662 0.001535
## sd.d 0.03318 0.02706 0.0004279 0.001681
##
## 2. Quantiles for each variable:
##
## 2.5% 25% 50% 75% 97.5%
## d.High.Moderate -0.1221050 -0.07454 -0.05168 -0.03040 0.019741
## d.High.Placebo -0.1342510 -0.08623 -0.06020 -0.03937 0.007528
## sd.d 0.0003187 0.01243 0.02794 0.04823 0.095717
##
## -- Model fit (residual deviance):
##
## Dbar pD DIC
## 48.16189 27.98140 76.14329
##
## 46 data points, ratio 1.047, I^2 = 7%5.3.4 収束の評価
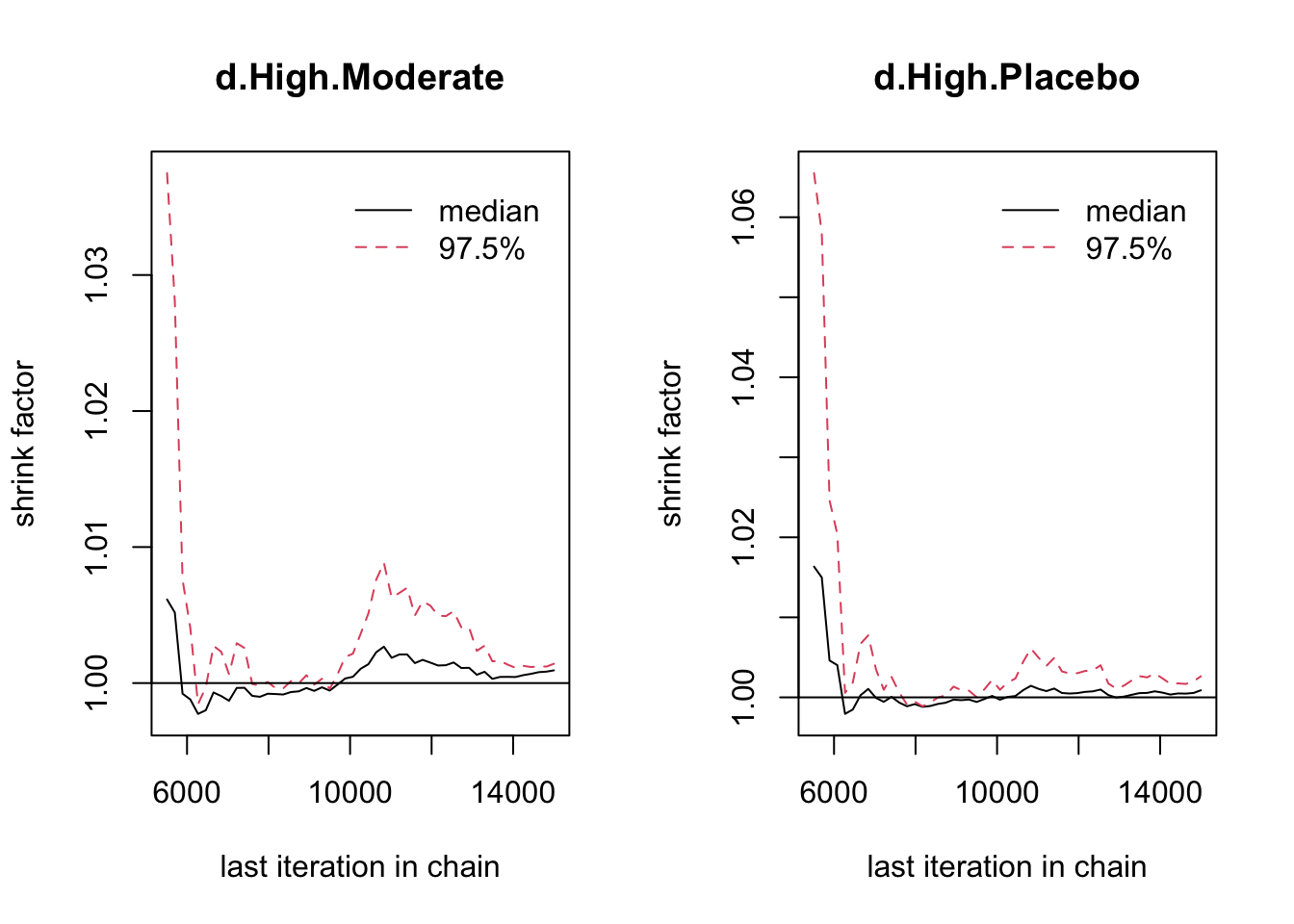
DIC は低い方が良い。Fixed が 74.75982、Random が75.98427。
## [1] 1.001424## [1] 1.0149651 に近い方が良い。
固定効果を採用。
5.3.6 一貫性の評価
エラーが出るため、検証中。
5.4 BUGSnet ベイジアン
BUGSnet では、arm-based データのみ対応している。
5.4.3 要約
TabBugsDavis <- net.tab(data = BugsDavis,
outcome = "X",
N = "Total",
type.outcome = "binomial",
time = NULL)
TabBugsDavis$intervention## # A tibble: 3 × 7
## treat n.studies n.events n.patients min.outcome max.outcome av.outcome
## <chr> <int> <int> <int> <dbl> <dbl> <dbl>
## 1 High 7 4654 28126 0.0181 0.435 0.165
## 2 Moderate 20 10946 62682 0 0.621 0.175
## 3 Placebo 19 9661 60013 0.000316 0.597 0.1615.4.5 モデル実行
n.adapt、n.inter、thin を色々試して、DIC 値を下げる。
## Compiling model graph
## Resolving undeclared variables
## Allocating nodes
## Graph information:
## Observed stochastic nodes: 46
## Unobserved stochastic nodes: 25
## Total graph size: 1003
##
## Initializing model## Compiling model graph
## Resolving undeclared variables
## Allocating nodes
## Graph information:
## Observed stochastic nodes: 46
## Unobserved stochastic nodes: 49
## Total graph size: 1040
##
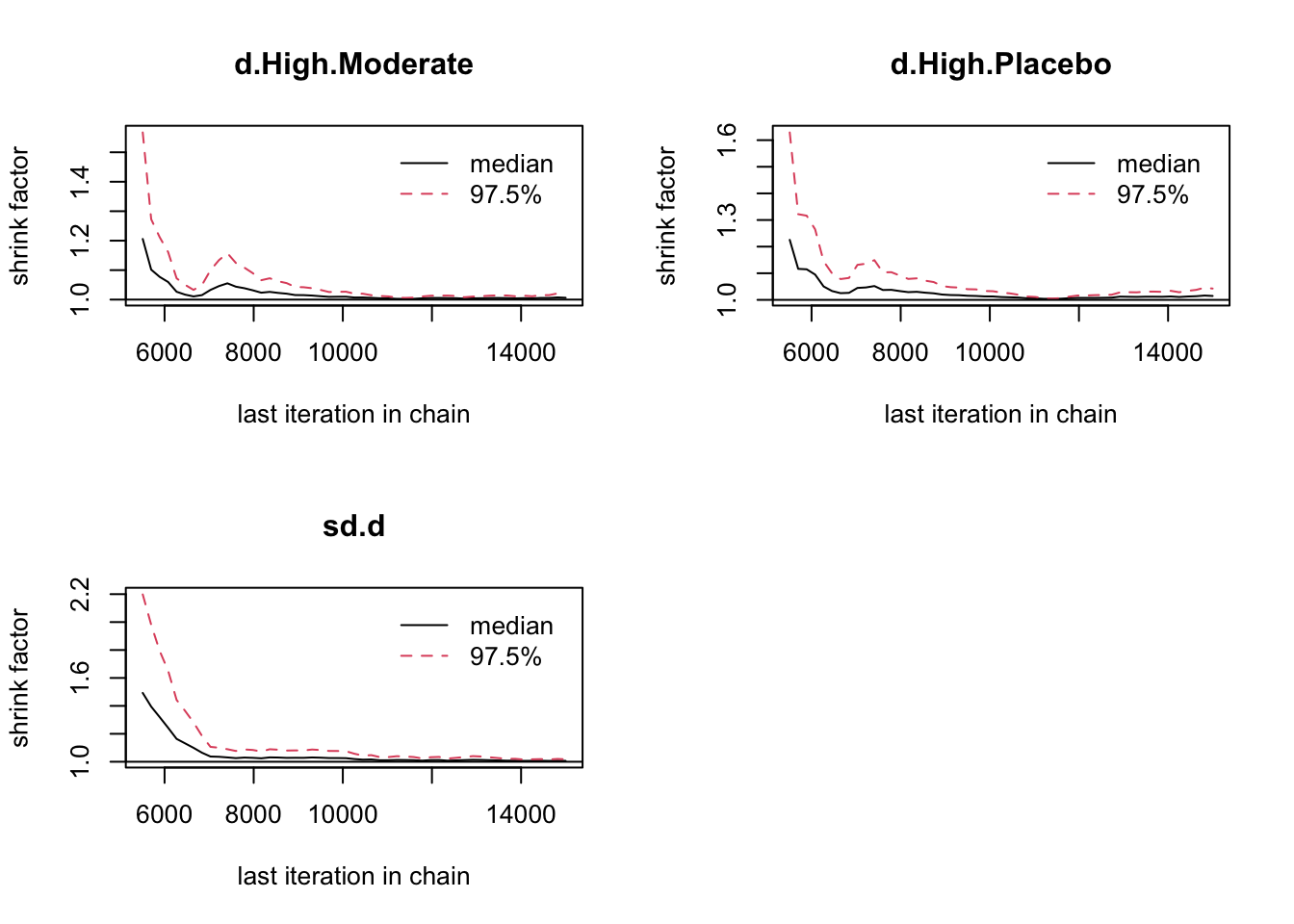
## Initializing model5.4.6 収束の評価
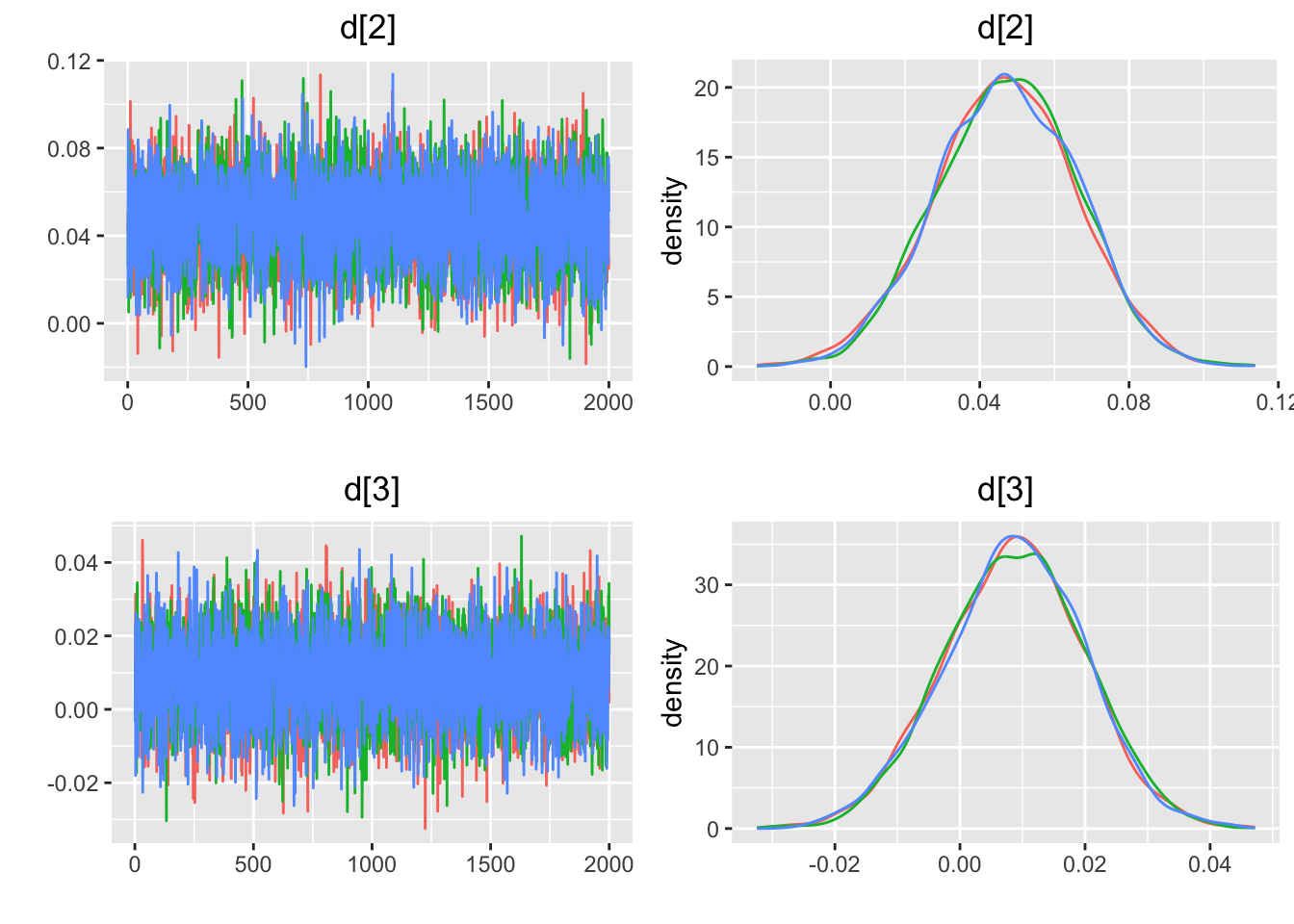
## $gelman.rubin
## $psrf
## Point est. Upper C.I.
## d[2] 1.000028 1.000726
## d[3] 1.000239 1.001535
##
## $mpsrf
## [1] 1.00032
##
## attr(,"class")
## [1] "gelman.rubin.results"
##
## $geweke
## $stats
## Chain 1 Chain 2 Chain 3
## d[2] -1.0241199 0.29017330 1.2089093
## d[3] 0.5759415 0.06487731 -0.2986586
##
## $frac1
## [1] 0.1
##
## $frac2
## [1] 0.5
##
## attr(,"class")
## [1] "geweke.results"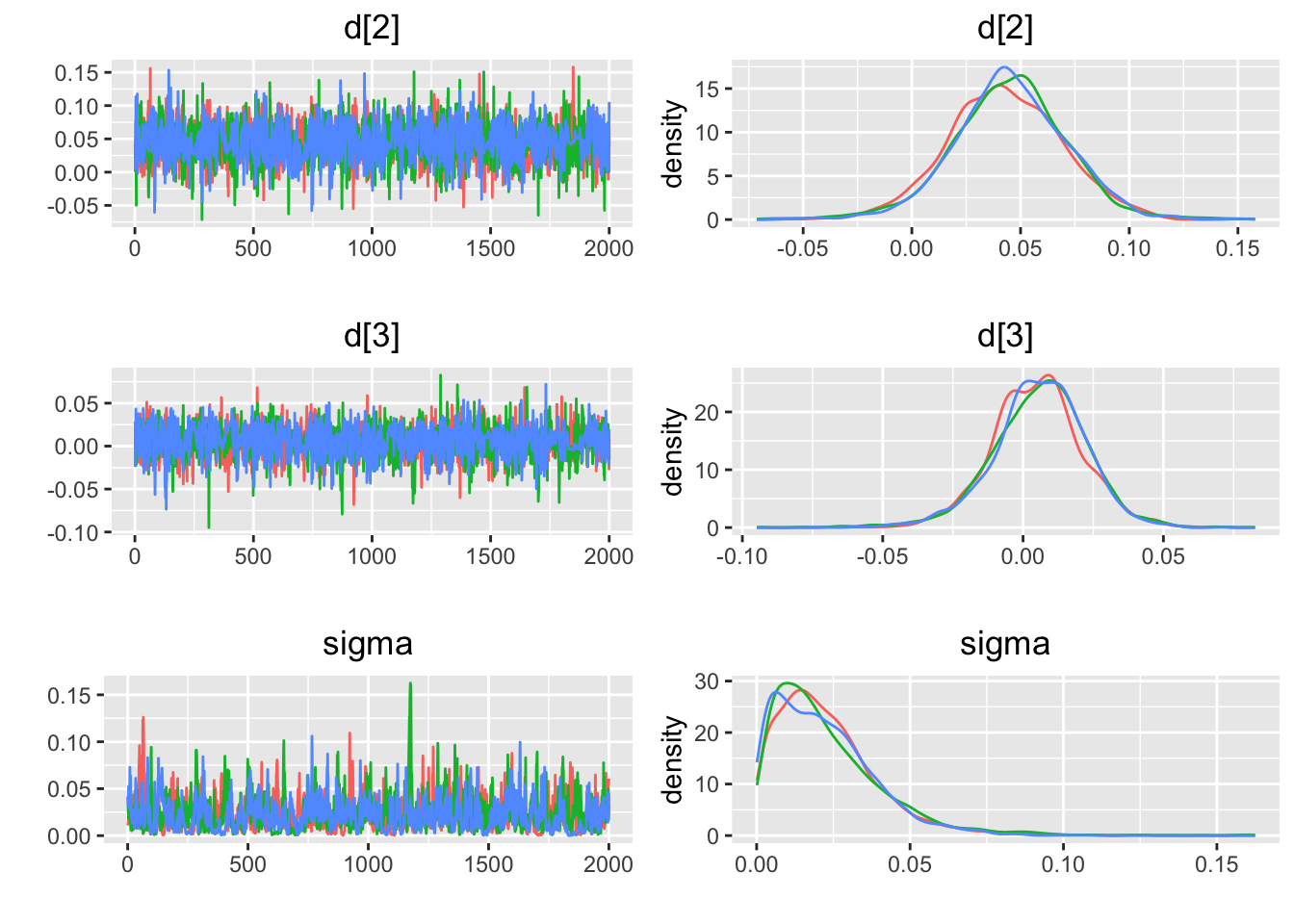
## $gelman.rubin
## $psrf
## Point est. Upper C.I.
## d[2] 1.001451 1.005226
## d[3] 1.002416 1.003517
## sigma 1.015574 1.025485
##
## $mpsrf
## [1] 1.003493
##
## attr(,"class")
## [1] "gelman.rubin.results"
##
## $geweke
## $stats
## Chain 1 Chain 2 Chain 3
## d[2] 0.1837099 0.5771300 -0.23362377
## d[3] 0.0948172 0.2085824 -0.04648464
## sigma 1.0791718 -1.4359398 1.50291837
##
## $frac1
## [1] 0.1
##
## $frac2
## [1] 0.5
##
## attr(,"class")
## [1] "geweke.results"5.4.7 適合度の評価
## $DIC
## [1] NaN
##
## $Dres
## [1] 48.59683
##
## $pD
## [1] NaN
##
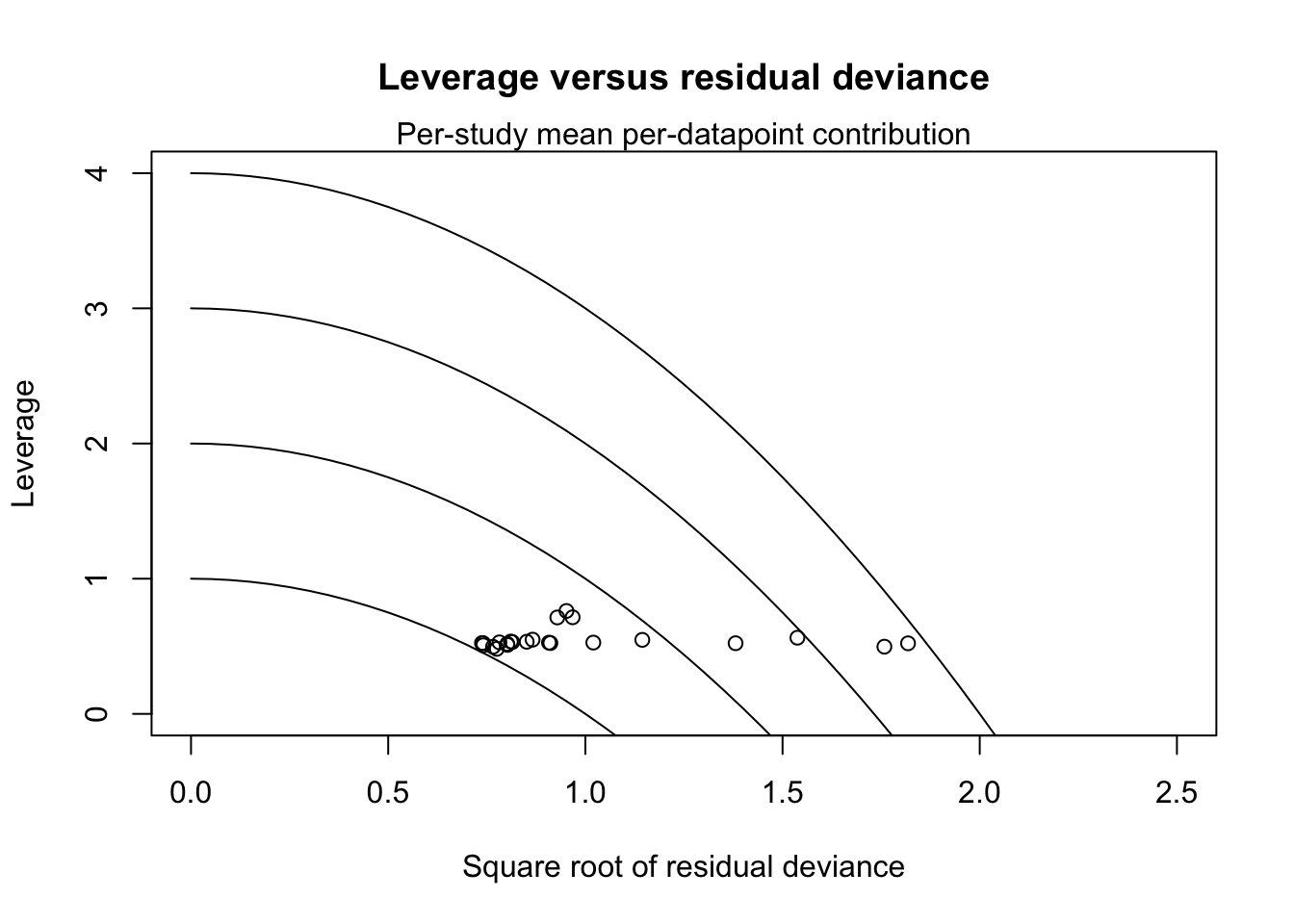
## $leverage
##
## 1 0.3917105 0.5135493 0.6601816 0.527994 0.492735 0.3446973 0.5229203 0.5080603
##
## 1 0.8223124 0.4955778 0.4653912 0.2130651 0.6613483 0.6453603 0.5338967
##
## 1 0.4641055 0.5692151 0.8257768 0.5357165 0.5269379 0.5421835 0.4982472
##
## 1 0.5087306 0.5463266 0.4273331 0.6578787 0.5226709 0.5246775 0.6440997
##
## 1 0.4951171 0.5216668 NaN 0.5356726 0.5094069 0.6381509 0.6488496 0.6697803
##
## 1 0.4277221 0.5215553 0.4595749 0.8355391 0.4891002 0.5389909 0.5269903
##
## 1 0.5581556 0.4851809
##
## $w
## r.1.1. r.2.1. r.3.1. r.4.1. r.5.1. r.6.1. r.7.1.
## -0.8890095 0.7864045 -1.3762191 0.7603438 -0.7930587 -1.8690288 1.3981543
## r.8.1. r.9.1. r.10.1. r.11.1. r.12.1. r.13.1. r.14.1.
## 0.7926714 1.2791974 -0.9077386 -0.7488842 -1.1608474 1.0287761 0.8554874
## r.15.1. r.16.1. r.17.1. r.18.1. r.19.1. r.20.1. r.21.1.
## 0.7506298 -0.8002997 0.9308799 0.9190619 1.0548751 -0.8289113 -0.7389336
## r.22.1. r.23.1. r.1.2. r.2.2. r.3.2. r.4.2. r.5.2.
## -0.8379113 0.7616019 0.7730933 -0.8218558 1.3868740 -0.7631022 0.7872248
## r.6.2. r.7.2. r.8.2. r.9.2. r.10.2. r.11.2. r.12.2.
## 1.6299092 -1.4233002 -0.8048225 -2.2256995 0.8695228 0.7187657 0.8548239
## r.13.2. r.14.2. r.15.2. r.16.2. r.17.2. r.18.2. r.19.2.
## -1.0239312 -0.8743759 -0.8375506 0.7683183 -0.9571942 -0.9272453 -1.1105402
## r.20.2. r.21.2. r.22.2. r.23.2.
## 0.8256853 0.7273149 0.8275154 -0.7829415
##
## $pmdev
## dev_a.1.1. dev_a.2.1. dev_a.3.1. dev_a.4.1. dev_a.5.1. dev_a.6.1.
## 0.7903378 0.6184321 1.8939791 0.5781227 0.6289420 3.4932687
## dev_a.7.1. dev_a.8.1. dev_a.9.1. dev_a.10.1. dev_a.11.1. dev_a.12.1.
## 1.9548356 0.6283280 1.6363460 0.8239894 0.5608275 1.3475667
## dev_a.13.1. dev_a.14.1. dev_a.15.1. dev_a.16.1. dev_a.17.1. dev_a.18.1.
## 1.0583803 0.7318587 0.5634450 0.6404796 0.8665374 0.8446748
## dev_a.19.1. dev_a.20.1. dev_a.21.1. dev_a.22.1. dev_a.23.1. dev_a.1.2.
## 1.1127615 0.6870940 0.5460228 0.7020953 0.5800374 0.5976733
## dev_a.2.2. dev_a.3.2. dev_a.4.2. dev_a.5.2. dev_a.6.2. dev_a.7.2.
## 0.6754470 1.9234195 0.5823250 0.6197228 2.6566040 2.0257834
## dev_a.8.2. dev_a.9.2. dev_a.10.2. dev_a.11.2. dev_a.12.2. dev_a.13.2.
## 0.6477392 4.9537381 0.7560700 0.5166242 0.7307239 1.0484352
## dev_a.14.2. dev_a.15.2. dev_a.16.2. dev_a.17.2. dev_a.18.2. dev_a.19.2.
## 0.7645332 0.7014910 0.5903130 0.9162207 0.8597838 1.2332995
## dev_a.20.2. dev_a.21.2. dev_a.22.2. dev_a.23.2.
## 0.6817562 0.5289870 0.6847817 0.6129975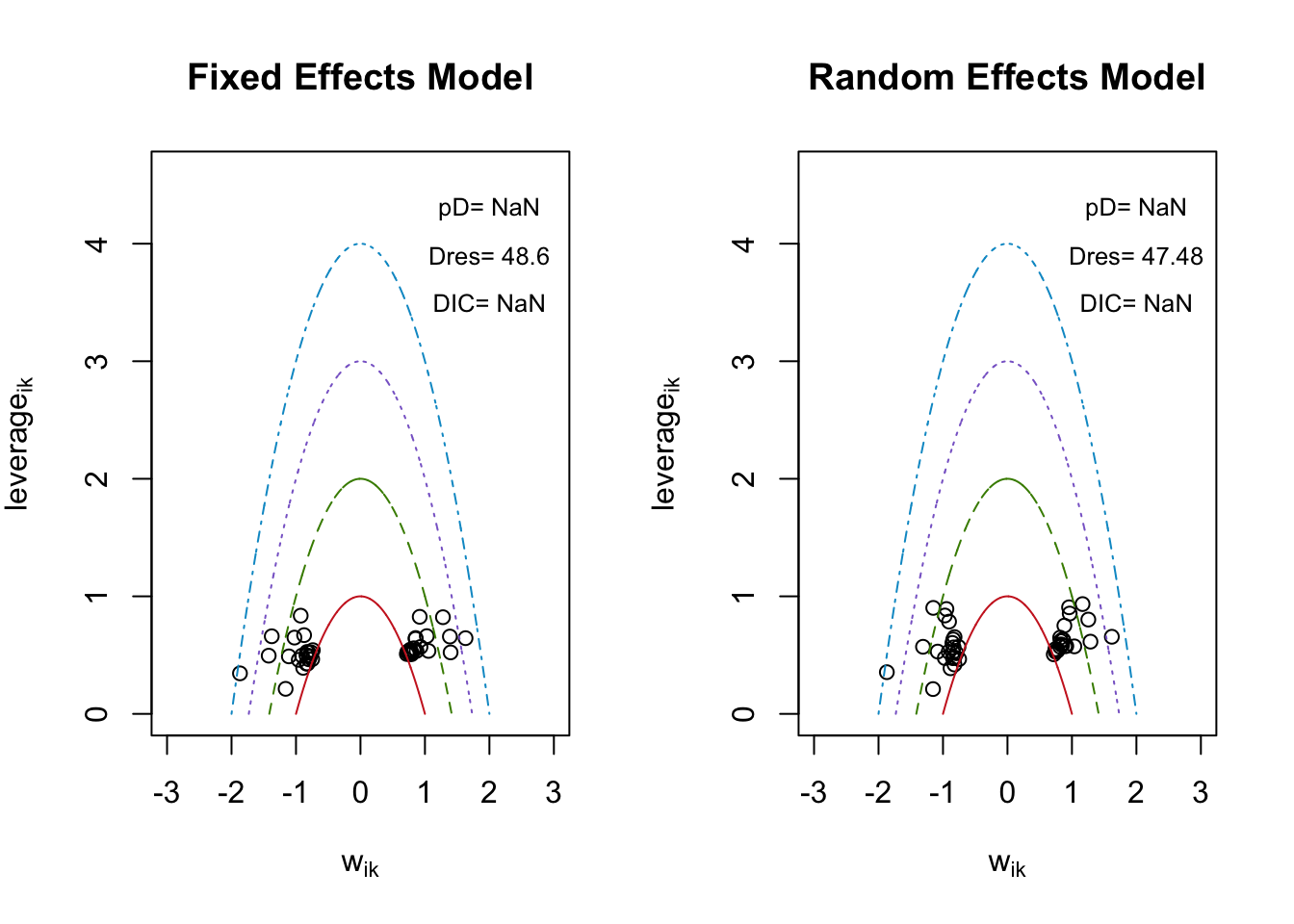
## $DIC
## [1] NaN
##
## $Dres
## [1] 47.48206
##
## $pD
## [1] NaN
##
## $leverage
##
## 1 0.3879883 0.5709878 0.9016669 0.6484318 0.5657538 0.3547065 0.6144591
##
## 1 0.6193026 0.8022221 0.5282019 0.4659938 0.2109391 0.8535578 0.7508483
##
## 1 0.5286571 0.4804632 0.5770053 0.9066433 0.5742166 0.6035965 0.5697343
##
## 1 0.524471 0.5421301 0.5434343 0.4736803 0.9340473 0.6531751 0.5910543
##
## 1 0.6560415 0.5707983 0.6331449 NaN 0.5766273 0.5064597 0.6329557 0.8369056
##
## 1 0.7850472 0.4198679 0.5365641 0.475001 0.8921608 0.5299677 0.627478 0.5505375
##
## 1 0.5854012 0.5160237
##
## $w
## r.1.1. r.2.1. r.3.1. r.4.1. r.5.1. r.6.1. r.7.1.
## -0.8830170 0.8182676 -1.1513447 0.8168469 -0.8351248 -1.8710162 1.2872817
## r.8.1. r.9.1. r.10.1. r.11.1. r.12.1. r.13.1. r.14.1.
## 0.8242121 1.2539918 -0.9121564 -0.7497071 -1.1549073 0.9652171 0.8827545
## r.15.1. r.16.1. r.17.1. r.18.1. r.19.1. r.20.1. r.21.1.
## 0.7474218 -0.8226919 0.9125323 0.9552475 1.0375318 -0.8545726 -0.7591116
## r.22.1. r.23.1. r.1.2. r.2.2. r.3.2. r.4.2. r.5.2.
## -0.8463772 0.7701794 0.7752967 -0.8465064 1.1645561 -0.8205691 0.8295536
## r.6.2. r.7.2. r.8.2. r.9.2. r.10.2. r.11.2. r.12.2.
## 1.6189623 -1.3101292 -0.8409299 -2.2355998 0.8914944 0.7180637 0.8557594
## r.13.2. r.14.2. r.15.2. r.16.2. r.17.2. r.18.2. r.19.2.
## -0.9704043 -0.9060763 -0.8220410 0.7733053 -0.9718819 -0.9468681 -1.0821366
## r.20.2. r.21.2. r.22.2. r.23.2.
## 0.8663692 0.7426384 0.8488024 -0.7932010
##
## $pmdev
## dev_a.1.1. dev_a.2.1. dev_a.3.1. dev_a.4.1. dev_a.5.1. dev_a.6.1.
## 0.7797191 0.6695619 1.3255947 0.6672389 0.6974334 3.5007017
## dev_a.7.1. dev_a.8.1. dev_a.9.1. dev_a.10.1. dev_a.11.1. dev_a.12.1.
## 1.6570941 0.6793256 1.5724954 0.8320293 0.5620608 1.3338109
## dev_a.13.1. dev_a.14.1. dev_a.15.1. dev_a.16.1. dev_a.17.1. dev_a.18.1.
## 0.9316440 0.7792556 0.5586393 0.6768220 0.8327152 0.9124977
## dev_a.19.1. dev_a.20.1. dev_a.21.1. dev_a.22.1. dev_a.23.1. dev_a.1.2.
## 1.0764723 0.7302943 0.5762504 0.7163544 0.5931762 0.6010850
## dev_a.2.2. dev_a.3.2. dev_a.4.2. dev_a.5.2. dev_a.6.2. dev_a.7.2.
## 0.7165730 1.3561909 0.6733336 0.6881592 2.6210390 1.7164384
## dev_a.8.2. dev_a.9.2. dev_a.10.2. dev_a.11.2. dev_a.12.2. dev_a.13.2.
## 0.7071631 4.9979063 0.7947623 0.5156155 0.7323241 0.9416846
## dev_a.14.2. dev_a.15.2. dev_a.16.2. dev_a.17.2. dev_a.18.2. dev_a.19.2.
## 0.8209742 0.6757514 0.5980010 0.9445543 0.8965593 1.1710196
## dev_a.20.2. dev_a.21.2. dev_a.22.2. dev_a.23.2.
## 0.7505955 0.5515119 0.7204655 0.6291679sD/DIC が計算されない。調査中。